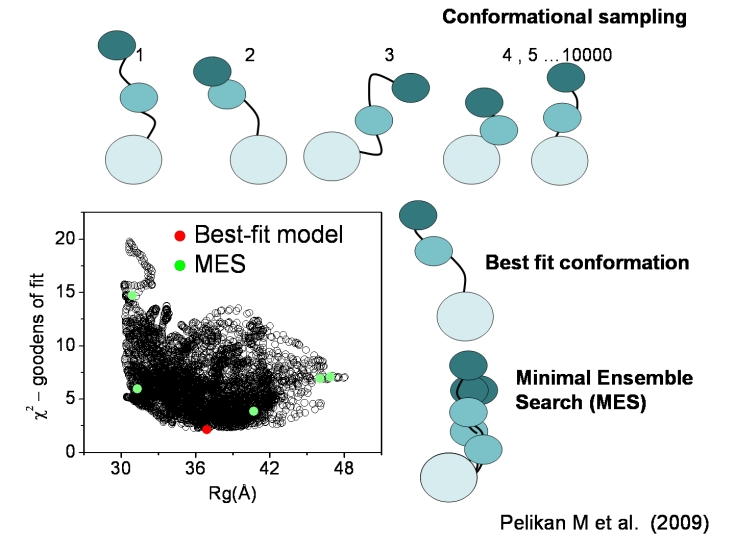
MES (Minimal Ensemble Search)
Version 1.x Copyright © Michal 2008
Written by: Martin Pelikan & Michal Hammel
Reference: Pelikan, M., Hura, G.L., and Hammel, M. (2009).
Structure and Flexibility within proteins as identified through small
angle X-ray scattering. General Physiology and Biophysics 28, 174-189
download program : MES
Hardware platforms: LINUX 32bit free available for academic use
USAGE on bl1231.als.lbl.gov
> ssh username@bl1231.als.lbl.gov # ask MH for access
> ssh crush !!!!!!!!
> cd to your directory with your scattering profiles
1. First you will need to clean head lines in the fit_files produced by FOXS. And have only one empty line at the top. please see the example for the *fit file. You may used FOXS web app to calculate fit_files.
2. Make filelist of the fit_files with the experimental_dat file at the top of the filelist ( experimental data containing q, I(q), SD example for experimental_dat )
Output:
Selected conformers and + weighting in %/100 will be printed at the end of the run.
avg_optimal is the MES -fit file with chi2 listed in the first line
for more details about mes usage see README