Structure of mammalian poly(ADP-ribose) glycohydrolase
Poly ADP-ribosylation regulates cellular processes such as genomic stability maintenance, transcription and cell death. The structure of rat poly(ADP-ribose) glycohydrolase has been determined using crystallographic data collected at the SIBYLS beamline, giving insights into the enzyme’s endoglycosidase activity and providing a basis for the development of therapeutic inhibitors.
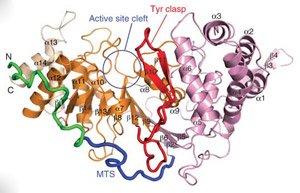
> Reversible post-translational modification by poly(ADP-ribose) (PAR) regulates chromatin structure, DNA repair and cell fate in response to genotoxic stress. PAR glycohydrolase (PARG) removes PAR chains from poly ADP-ribosylated proteins to restore protein function and release oligo(ADP-ribose) chains to signal damage. Here we report crystal structures of mammalian PARG and its complex with a substrate mimic that reveal an open substrate-binding site and a unique ‘tyrosine clasp’ enabling endoglycosidic cleavage of branched PAR chains.