Nanogold labels used to characterize DNA bending in mismatch repair
Greg Hura and colleagues developed and applied nanogold labels for DNA complexes with proteins examined by small angle X-ray scattering (SAXS) to follow DNA conformations acting in error detection by the mismatch repair (MMR) system in solution. This technique can examine short or long pieces of DNA and in most solution conditions, including those closest to cellular environments. This technique is expected to be useful for many biologically important systems involving DNA complexes and conformations. In this manuscript the authors reveal DNA bending followed by straightening by the repair protein MutS at the site of a mismatch as a suitable mechanism for error detection and signaling needed to avoid mutations and cancers and to control microbial stability and evolution in response to environmental stress.

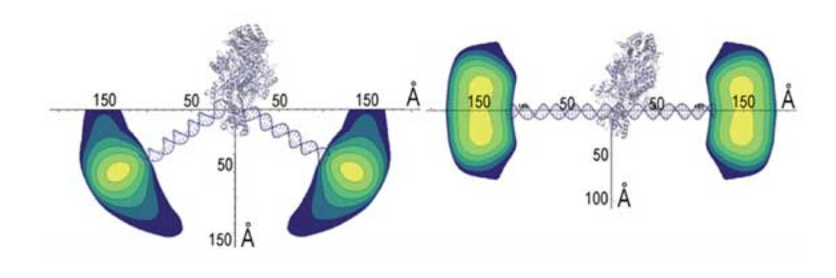
Mismatch DNA bending by MutS and straightening in the presence of ATP. Contour plots of the distribution of DNA ends are visualized by placing the structural information from the crystal structure of MutS/DNA on the same scale as the distance and pop- ulation information from the *P(Dij)* distributions. The *P(Dij)* distributions from 71-bp DNA in the presence of MutS (**left**) and the presence of MutS and excess ATP (**right**) set contour levels. The widest part of the distribution is the width of the gold nanocrystal. DNA of the crystal structure has been ex- tended to 71 bp for the MutS/DNA complex and replaced by straight DNA for the ATP model.
For all the details please check out the full manuscript: