Exploring the Repeat Protein Universe through Computational Protein Design
The facilities and staff at the SYBILS beamline contributed to this breakthrough study exploring the extent to which naturally occurring proteins sample the space of folded structures accessible to the polypeptide chain.

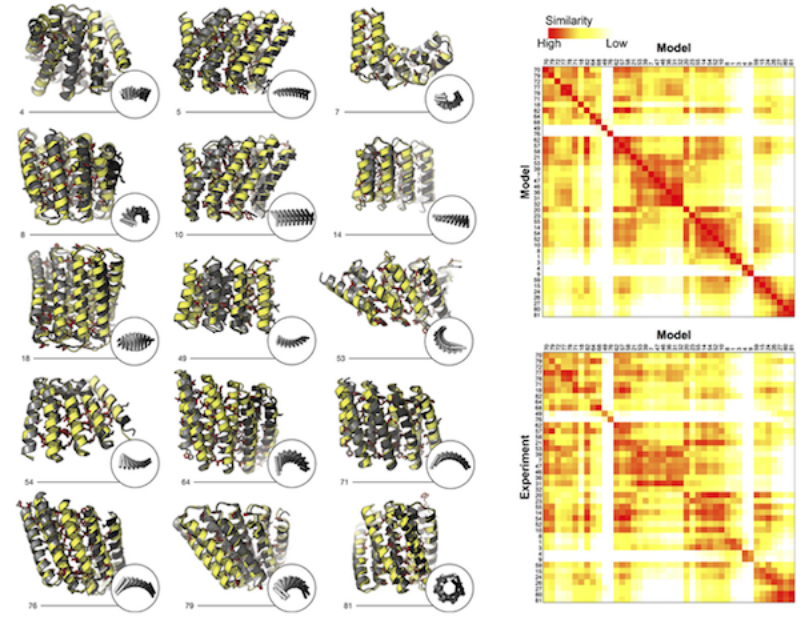
Naturally occurring proteins–chains of amino acids that fold into functional, three-dimensional shapes–are believed to represent just a small fraction of the universe of all possible permutations of amino-acid sequences and folds. How can we begin to systematically sift through those permutations to find and engineer from scratch (de novo) proteins with the characteristics desired for medical, environmental, and industrial purposes? To address this question, a team led by researchers from the Institute for Protein Design at the University of Washington have published a landmark study that used both protein crystallography and small-angle x-ray scattering (SAXS) at the ALS to validate the computationally designed structures of novel proteins with repeated motifs. The results show that the protein-folding universe is far larger than realized, opening up a wide array of new possibilities for biomolecular engineering.