Predicting RNA structure and dynamics with deep learning and solution scattering
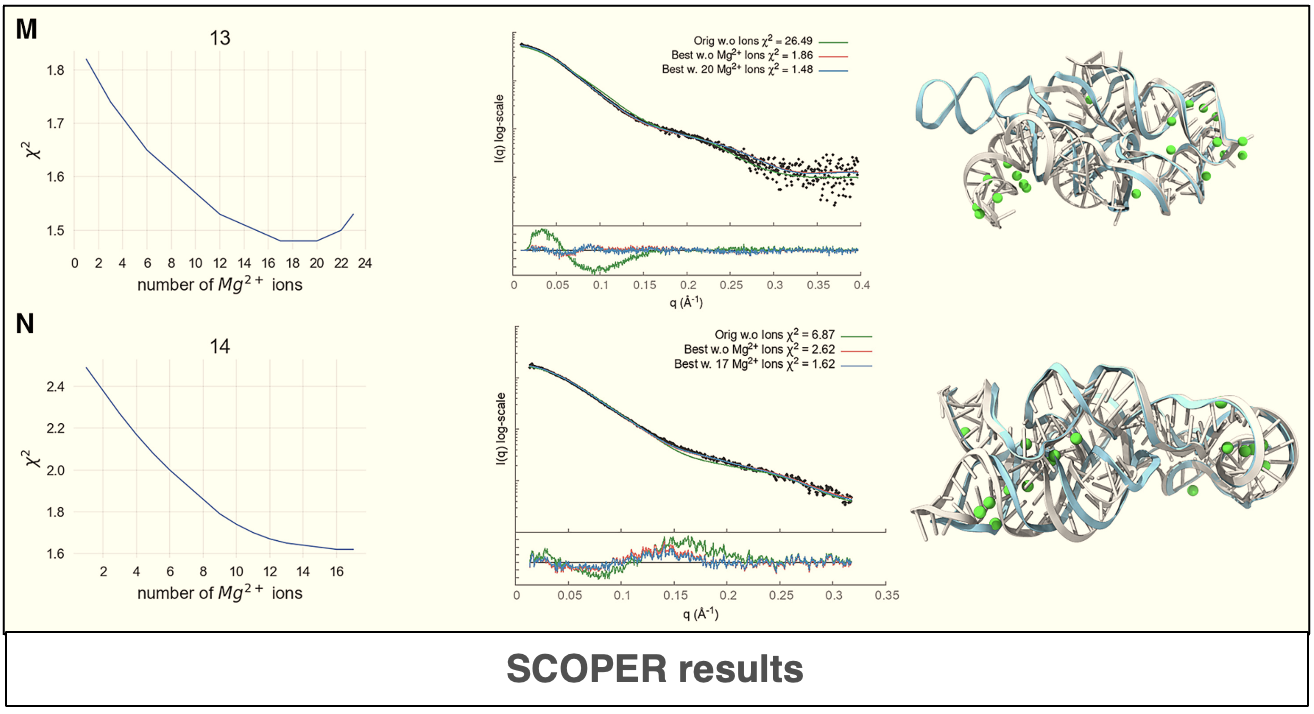
The use of deep-learning and statistical methods plays a significant role in the prediction of accurate structure and atomistics RNA models. In this paper, the authors describe new a deep-learning tool they developed called Scoper, which is capable of using experimental SAXS data from the SIBYLS beam line to determine the most likely 3-dimensional conformation of an RNA molecule as well as predicting the number and positions of Magnesium ions. The Scoper pipeline has been implemented using the existing BilboMD webapp to help understand the behavior of RNA in solution which is critical for deciphering its biological functions.
Predicting Magnesium positions and understanding the behavior of RNA in solution can be challenging. This study introduces an innovative tool that offers a more reliable approach for validating and refining RNA stuctures against experimental SAXS data. The authors provide a link to the web server, BilboMD, that runs the Scoper pipeline.