Formation of repressive complex in the mammalian circadian clock is demoted by the secondary pocket of CRY1
In this paper, the authors show that CRY1, a protein coding gene that activates circadian gene expression and metabolic states and circadian oscillators, binds directly to the PAS domain core of CLOCK:BMAL1. Precise control of CLOCK:BMAL1 activity by coactivators and repressors establishes the ~24 hr periodicity of gene expression. Integrative modeling and solution X-ray scattering studies (conducted at the SIBYLS beamline 12.3.1) irrefutably position a key loop of the CLOCK PAS-B domain in the secondary pocket of CRY1, analogous to the antenna chromophore-binding pocket of photolyase. This study is significant for understanding the clock mechanism as fundamental for the development and application of therapies for circadian-related disorders.

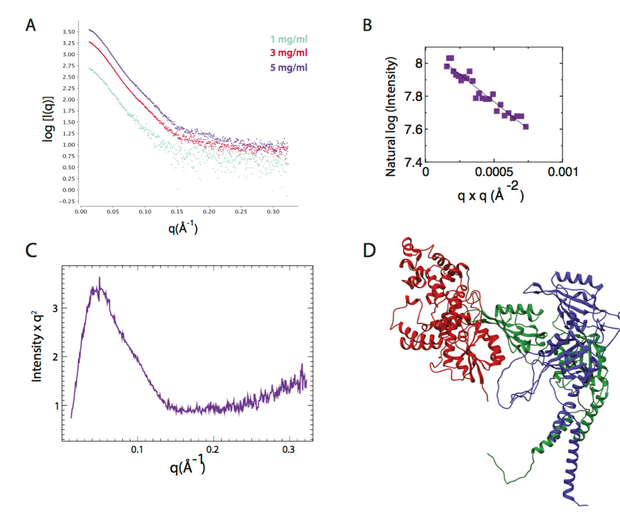
SAXS profile of CRY1:CLOCK:BMAL1 repressive complex.
(A) Scattering traces of CRY1:CLOCK:BMAL1 ternary complex (CCB) at different con- centrations are shown. These scattering plots were merged to generate the dataset as the input for FoXSDock. (B) Guinier analysis of CCB shows little or no aggregation of sample. SAXS-calculated molecular weight of the ternary complex is 113 kDa. (C) Kratky plot shows the CCB complex indicates a folded mass with an elongated shape. (D) PDB of FoXSDock HADDOCK driven model that is among the top 20 nearly degenerate docking structures, χ = 2.74.
Michael AK, Fribourgh L, Chelliah Y, Sandate C, Hura GL, Schneidman-Duhovny, Tripathi SM, Takahashi JS, Partch CL [“Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1”](http://www.pnas.org/content/early/2017/01/30/1615310114.full.pdf?sid=22e8f8bf-7098-4c1c-a81a-e26fb35c50bf) PNAS 2017 Jan 31, doi:10.1073/pnas.1615310114